Winnow
Confidence calibration and FDR control for de novo peptide sequencing
Explore the docs »
Report bug
·
Request feature
Table of Contents
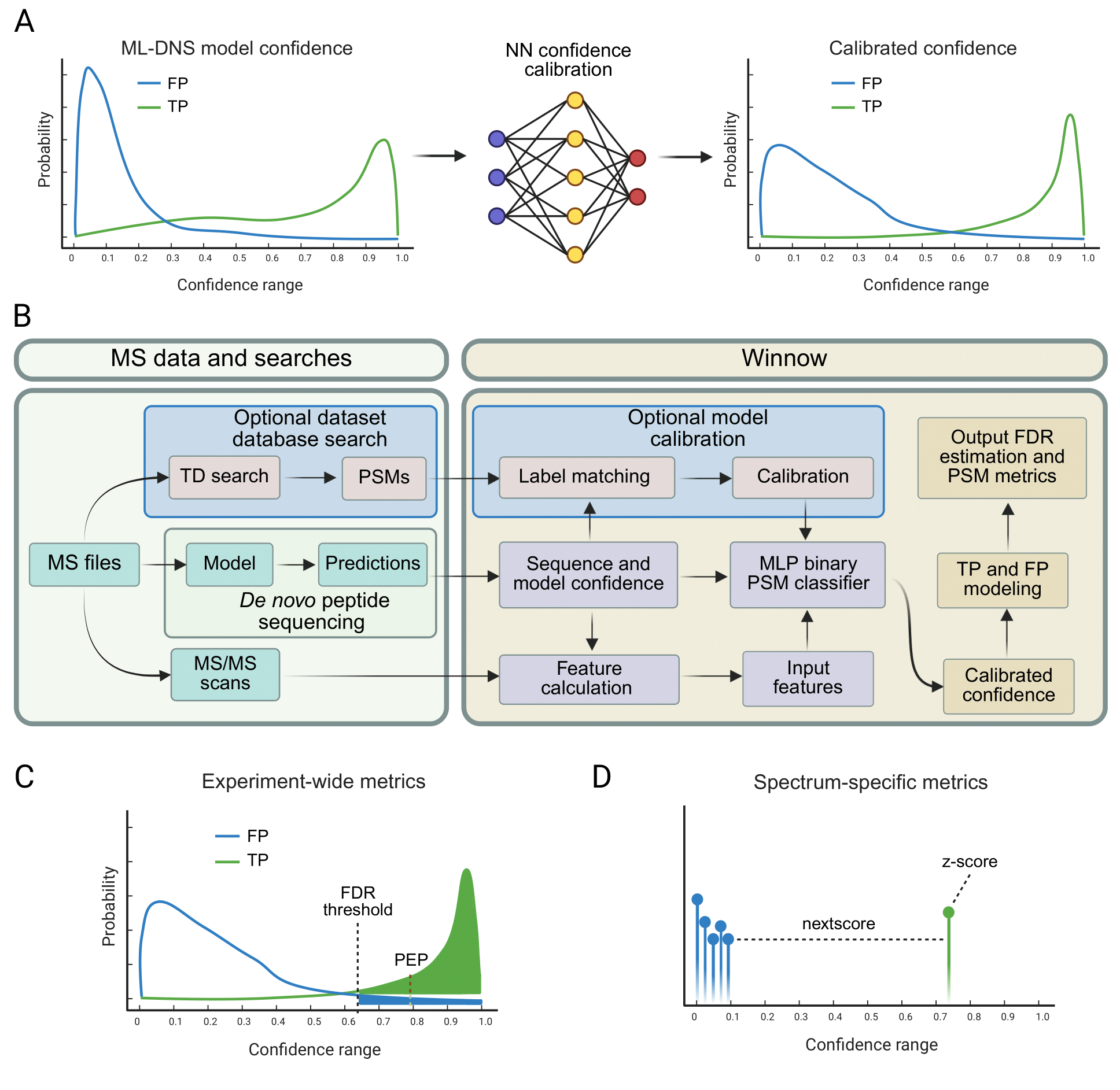
Winnow workflow for confidence calibration and FDR control in de novo peptide sequencing
About the project¶
In bottom-up proteomics workflows, peptide sequencing—matching an MS2 spectrum to a peptide—is just the first step. The resulting peptide-spectrum matches (PSMs) often contain many incorrect identifications, which can negatively impact downstream tasks like protein assembly.
To mitigate this, intermediate steps are introduced to:
- Assign confidence scores to PSMs that better correlate with correctness.
- Estimate and control the false discovery rate (FDR) by filtering identifications based on confidence scores.
For database search-based peptide sequencing, PSM rescoring and target-decoy competition (TDC) are standard approaches, supported by an extensive ecosystem of tools. However, de novo peptide sequencing lacks standardised methods for these tasks.
winnow aims to fill this gap by implementing the calibrate-estimate framework for FDR estimation. Unlike TDC, this approach is directly applicable to de novo sequencing models. Additionally, its calibration step naturally incorporates common confidence rescoring workflows as part of FDR estimation.
winnow provides both a CLI and a Python package, offering flexibility in performing confidence calibration and FDR estimation.
Installation¶
winnow is available as a Python package and can be installed using pip or a pip-compatible command (e.g., uv pip install):
Quick Start¶
Get started with winnow in minutes using the included sample data:
# Generate sample data (if not already present)
make sample-data
# Train a calibrator on the sample data using default settings
make train-sample
# Run prediction with the trained model without filtering on an FDR threshold
make predict-sample
Note: The sample data is minimal (20 spectra) and intended for testing only. The make commands shown above are configured for the sample data with adjusted settings (e.g., relaxed FDR threshold). For your own datasets, use the winnow commands outlined below.
Usage¶
winnow supports two usage modes:
- A command-line interface (CLI) with sensible defaults and multiple FDR estimation methods.
- A configurable and extensible Python package for advanced users.
CLI¶
Installing winnow provides the winnow command with two sub-commands:
winnow train– Performs confidence calibration on a dataset of annotated PSMs, outputting the fitted model checkpoint.winnow predict– Performs confidence calibration using a fitted model checkpoint (defaults to a pretrained general model from HuggingFace), estimates and controls FDR using the calibrated confidence scores.
By default, winnow predict uses a pretrained general model (InstaDeepAI/winnow-general-model) hosted on HuggingFace Hub, allowing you to get started immediately without training. You can also specify custom HuggingFace models or use locally trained models.
Winnow uses Hydra for flexible, hierarchical configuration management. All parameters can be configured via YAML files or overridden on the command line:
# Quick start with defaults
winnow predict
# Override specific parameters
winnow predict fdr_control.fdr_threshold=0.01
# Specify different data source and dataset paths
winnow predict data_loader=mztab dataset.spectrum_path_or_directory=data/spectra.parquet dataset.predictions_path=data/preds.mztab
Refer to the CLI Guide and Configuration Guide for details on usage and configuration options.
Package¶
The winnow package is organised into three sub-modules:
winnow.datasets– Handles data loading and saving, including theCalibrationDatasetclass for mapping peptide sequencing output formats.winnow.calibration– Implements confidence calibration. Key components include:ProbabilityCalibrator(defines the calibration model)CalibrationFeature(an extensible interface for defining calibration features)
winnow.fdr– Implements FDR estimation methods:DatabaseGroundedFDRControl(for database-grounded FDR control)NonParametricFDRControl(uses a non-parametric and label-free method for FDR estimation)
For an example, check out the example notebook.
Contributing¶
Contributions are what make the open-source community such an amazing place to learn, inspire and create, and we welcome your support! Any contributions you make are greatly appreciated.
If you have ideas for enhancements, you can:
- Fork the repository and submit a pull request.
- Open an issue and tag it with "enhancement".
Contribution process¶
- Fork the repository.
- Create a feature branch (
git checkout -b feat-amazing-feature). - Commit your changes (
git commit -m 'feat: add some amazing feature'). - Push to your branch (
git push origin feat-amazing-feature).
Don't forget to give the project a star! Thanks again!
BibTeX entry and citation information¶
If you use winnow in your research, please cite the following preprint:
@article{mabona2025novopeptidesequencingrescoring,
title={De novo peptide sequencing rescoring and FDR estimation with Winnow},
author={Amandla Mabona and Jemma Daniel and Henrik Servais Janssen Knudsen
and Rachel Catzel and Kevin Michael Eloff and Erwin M. Schoof and Nicolas Lopez
Carranza and Timothy P. Jenkins and Jeroen Van Goey and Konstantinos Kalogeropoulos},
year={2025},
eprint={2509.24952},
archivePrefix={arXiv},
primaryClass={q-bio.QM},
url={https://arxiv.org/abs/2509.24952},
}




