Quick overview
This tutorial will guide you through the first steps of using InstaNovo, from installation to making your first predictions.
What is de novo peptide sequencing?
In proteomics, scientists study proteins, which are large, complex molecules made up of smaller units called amino acids. To understand a protein's function, we often need to know its amino acid sequence.
Mass spectrometry is a technique used to measure the mass-to-charge ratio of ions. In a typical proteomics experiment, proteins are broken down into smaller pieces called peptides. These peptides are then analyzed by a mass spectrometer, which generates a spectrum of peaks. Each peak corresponds to a fragment of the peptide.
De novo peptide sequencing is the process of determining the amino acid sequence of a peptide directly from its tandem mass spectrum (MS/MS) without relying on a sequence database. This is where InstaNovo comes in.
Installation
To use InstaNovo, you first need to install the Python package. You can do this using pip.
If you have access to an NVIDIA GPU, we recommend installing InstaNovo with the GPU version of PyTorch for better performance:
If you are on a machine without a GPU, or on macOS, you can install the CPU-only version:
InstaNovo now has support for Metal Performance Shaders (MPS) for Apple silicon devices. If you would like to use InstaNovo with MPS, please set 'mps' to True in the configuration files (instanovo/configs/) and set the environment variable:
This allows the CPU to be used for functionality not yet supported on MPS. Note torch<2.8.0 is required if running on MPS.
For more details on installation for development, see the development guide.
Making your first prediction
Once InstaNovo is installed, you can use the command-line interface (CLI) to make predictions.
Let's try predicting the peptide sequence from a sample spectrum file. InstaNovo comes with a sample data file (spectra.mgf) for this purpose.
The following command runs prediction with the base InstaNovo model and iteratively refines the results with the InstaNovo+ model:
For a full overview of how to make predictions with our different models, see our predicting how-to guide.
Understanding the output
The instanovo predict command in the previous step created a file named predictions.csv with the predicted peptide sequences:
experiment_name,scan_number,spectrum_id,precursor_mz,precursor_charge,prediction_id,predictions,log_probs,token_log_probs,group,instanovo_predictions,instanovo_log_probabilities,instanovo_token_log_probabilities,instanovo_predictions_beam_0,instanovo_log_probabilities_beam_0,instanovo_token_log_probabilities_beam_0,instanovo_predictions_beam_1,instanovo_log_probabilities_beam_1,instanovo_token_log_probabilities_beam_1,instanovo_predictions_beam_2,instanovo_log_probabilities_beam_2,instanovo_token_log_probabilities_beam_2,instanovo_predictions_beam_3,instanovo_log_probabilities_beam_3,instanovo_token_log_probabilities_beam_3,instanovo_predictions_beam_4,instanovo_log_probabilities_beam_4,instanovo_token_log_probabilities_beam_4,diffusion_predictions,diffusion_log_probabilities,diffusion_token_log_probabilities,diffusion_unrefined_predictions,diffusion_predictions_beam_0,diffusion_log_probabilities_beam_0,diffusion_predictions_beam_1,diffusion_log_probabilities_beam_1,diffusion_predictions_beam_2,diffusion_log_probabilities_beam_2,diffusion_predictions_beam_3,diffusion_log_probabilities_beam_3,diffusion_predictions_beam_4,diffusion_log_probabilities_beam_4,predictions_tokenised,delta_mass_ppm
spectra,0,spectra:0,451.25348,2,0,LAHYNKR,-0.04958329349756241,,no_group,"['L', 'A', 'H', 'Y', 'N', 'K', 'R']",-1.4490094184875488,[0],LAHYNKR,-1.4490094184875488,[0],LAHYNKR,-1.7640595436096191,[0],LAHYNKR,-1.7640595436096191,[0],LAHYNKR,-1.7640595436096191,[0],LAHYNKR,-1.7640595436096191,[0],"['L', 'A', 'H', 'Y', 'N', 'K', 'R']",-0.04958329349756241,,"['L', 'A', 'H', 'Y', 'N', 'K', 'R']","['L', 'A', 'H', 'Y', 'N', 'K', 'R']",-0.260211318731308,"['L', 'A', 'H', 'Y', 'N', 'K', 'R']",-0.5505515336990356,"['L', 'A', 'H', 'Y', 'N', 'K', 'R']",-0.08841510117053986,"['L', 'A', 'H', 'Y', 'N', 'K', 'R']",-0.04958329349756241,"['I', 'A', 'H', 'Y', 'N', 'K', 'R']",-0.1989564299583435,"L, A, H, Y, N, K, R",0.6781111138830191
This output CSV file contains several columns. Here are some of the most important ones:
| Column | Description |
|---|---|
scan_number |
The scan number of the spectrum in the input file. |
precursor_mz |
The mass-to-charge ratio of the precursor ion. |
precursor_charge |
The charge of the precursor ion. |
diffusion_predictions |
The peptide sequence predicted by InstaNovo+. |
transformer_predictions |
The peptide sequence predicted by the base InstaNovo model. |
log_probs |
The log probability of the prediction. Higher (less negative) values indicate greater model confidence in the predicted output. |
For a full description of the output, see the prediction output reference.
Evaluating performance
If your input data is annotated with the true peptide sequences, you can evaluate InstaNovo's performance.
Use the --evaluation flag to enable evaluation mode:
This will add a targets column to the output file with the ground truth sequences and will print performance metrics to the console.
For more background regarding the metrics we use for evaluation, see our explanation on performance metrics.
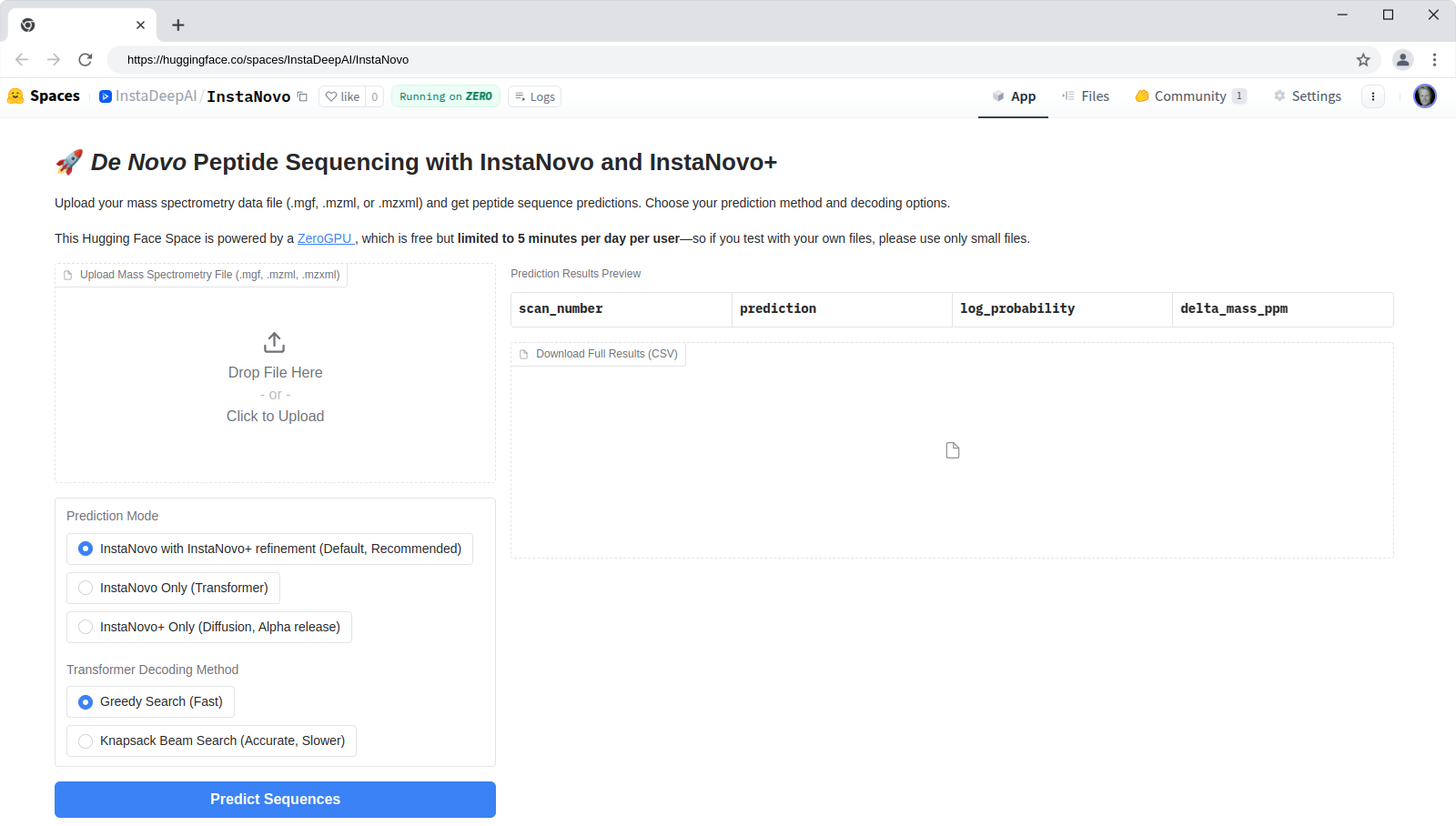
HuggingFace Space
InstaNovo is available as a HuggingFace Space at
hf.co/spaces/InstaDeepAI/InstaNovo for quick
testing and evaluation. You can upload your own spectra files in .mgf, .mzml, or .mzxml format
and run de novo predictions. The results will be displayed in a table format, and you can download
the predictions as a CSV file. The HuggingFace Space is powered by the InstaNovo model and the
InstaNovo+ model for iterative refinement.
Next steps
Now that you've made your first predictions, you can explore more of what InstaNovo has to offer:
- Learn how to make predictions with different models and settings in the predicting how-to guide.
- Find out how to train your own models in the training how-to guide.
- Dive deeper into the concepts behind InstaNovo in the explanation section.